DNA translation : is the process which can be seen as the decoding of instructions for making proteins, it is part of the process of gene expression.
The process by which DNA is copied to RNA is called transcription, and that by which RNA is used to produce proteins is called translation.
Translation usually happens in the cytoplasm where the ribosomes are located.
Translation proceeds in three phases: initiation, elongation and termination.
Initiation:
1- factors IF–1 and IF–3, bind to the 30 S subunit
2- Another factor,
IF–2, binds as a complex with GTP
3- This allows the subunit to associate with the mRNA and
makes it possible for a special tRNA to bind to the start codon ………………………… (IF released)
4- In prokaryotes,
this starter tRNA carries the substituted amino acid Nformylmethionine
(fMet). In eukaryotes, it carries an unsubstituted
methionine.
·
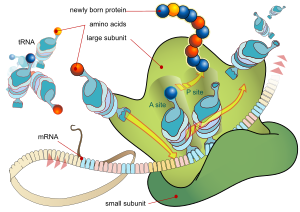
In the 70S initiation
complex, formylmethionine tRNA is initially located at a binding site known as
the peptidyl site (P).
·
Acceptor site (A), is not
yet occupied during this phase of translation.
·
Sometimes, a third tRNA
binding site is defined as an exit site (E).
The anticodon of another tRNA binds to the next mRNA
codon, bringing the 2nd amino acid to be placed in the protein.
As each anticodon & codon bind together a peptide
bond forms between the two amino acids.
After translation has been initiated , the peptide chain
is extended by the addition of further amino acid residues (elongation) until
the ribosome reaches a stop codon on the mRNA and the process is interrupted
(termination).
Elongation:
Elongation can be divided into four phases:
[1] Binding of aminoacyl tRNA.
1a . First, the peptidyl site (P) of the ribosome is
occupied by a tRNA that carries at its 3 end the complete peptide chain formed
up .
-A second tRNA, loaded with the next amino acid, then binds
via its complementary anticodon to the mRNA codon exposed at the acceptor site.
1b. The tRNA binds as a complex with a GTP containing
protein, the elongation factor Tu (EF–Tu) . after the bound GTP has been
hydrolyzed to GDP and phosphate that EF–Tu dissociates again.
-As the binding of the tRNA to the mRNA is still loose
before this, GTP hydrolysis acts as a delaying factor, making it possible to
check whether the correct tRNA has been bound.
-A further protein, the elongation factor Ts (EF-Ts), later
catalyzes the exchange of GDP for GTP and in this way regenerates the EF–Tu GTP
complex. EF-Tu is related to the G proteins involved in signal transduction.
[2] Synthesis of the peptide bond takes
place in the next step. Ribosomal peptidyl transferase
catalyzes (without consumption of ATP or GTP) the transfer of the peptide chain
from the tRNA at the P site to the NH2 group of the amino acid residue of the
tRNA at the A site.
The ribosome’speptidylt ransferase activity is not located in one of the
ribosomal proteins, but in the 28 S rRNA. Catalytically active RNAs of this
type are known as ribozymes
[3]. free tRNA at the P site dissociates and another
GTP–containing elongation factor (EF- GTP) binds to the ribosome
Hydrolysis of the GTP in this factor provides the energy
[4]-translocation of the ribosome.
The uncharged Val-tRNA then dissociates from the E site. The
ribosome is now ready for the next elongation cycle.
termination:
There are three termination codons, employed at the end of a
protein-coding sequence in mRNAand there are not tRNAs recognize these
codons.these codons are :
UAA
UAG
UGA
Thus, in the place of these tRNAs, one of several proteins, called release factors, binds and facilitates release of the mRNA from the ribosome and subsequent dissociation of the ribosome.
Eukaryotic and Prokaryotic Translation
UAA
UAG
UGA
Thus, in the place of these tRNAs, one of several proteins, called release factors, binds and facilitates release of the mRNA from the ribosome and subsequent dissociation of the ribosome.
Eukaryotic and Prokaryotic Translation
The translation process is very similar in prokaryotes and eukaryotes.
Although different elongation, initiation, and termination factors are used,
the genetic code is generally identical. In bacteria,
transcription and translation take place together, and mRNAs are
relatively short-lived. In eukaryotes, however, mRNAs have highly variable
half-lives, are subject to modifications and must exit the nucleus to be
translated; these multiple steps offer additional opportunities to regulate levels
of protein production, and thereby fine-tune gene expression.
Energy requirements in protein synthesis are high.
= two energy-rich bonds per amino acid activation (ATP AMP + PP)
-initiation requires one GTP per chain.
GTPs are consumed per elongation cycle.
Termination requires one GTP.









0 التعليقات:
Post a Comment